Transcription, the process of creating RNA from a DNA template, is fundamental to life. But Where Does Transcription Happen within the bustling environment of a cell, especially when another critical process, DNA replication, is underway? New research sheds light on this intricate coordination, revealing that transcription not only occurs during DNA replication but also faces unique challenges at specific genomic locations, impacting genome stability.
The Transcription-Replication Balancing Act
DNA replication and transcription both rely on DNA as their substrate. This shared dependency can lead to conflicts, potentially causing genome instability and health issues. Imagine two trains needing to use the same track simultaneously – coordination is key to avoid collisions. In cells, these “collisions” between transcription and replication machinery can hinder DNA duplication and increase the risk of DNA damage.
Scientists have long debated whether transcription and replication are spatially or temporally separated within the cell. Some studies suggested they occur in distinct areas, while others pointed to overlaps. Adding to the complexity, transcription can happen at any stage of the cell cycle, whereas DNA replication is primarily confined to the S phase. RNA polymerase II (RNAPII), the workhorse of protein-coding gene transcription, is even known to transcribe genes involved in replication itself during S phase. This raises the critical question: how do these essential processes coexist and influence each other within the cell?
To delve deeper into this interplay, researchers conducted a detailed investigation into the spatial and temporal relationship between transcription and replication throughout the S phase of the cell cycle.
Transcription Persists During DNA Replication, But Faces Roadblocks
Using advanced genomic techniques, scientists discovered that transcription continues to occur even as DNA replication progresses. However, this active transcription has notable consequences, particularly at transcription start sites (TSSs) – the regions where genes begin to be transcribed.
 Genomic view of BrdU-seq and Chr-RNA-seq
Genomic view of BrdU-seq and Chr-RNA-seq
One key finding was that TSSs are not fully replicated at the same pace as the surrounding DNA regions. These crucial gene initiation points tend to be under-replicated during the main DNA replication phase (S phase), lagging behind in the duplication process.
Intriguingly, the research revealed that the completion of DNA replication at TSSs often gets pushed to a later stage – specifically, when cells enter mitosis. Mitosis is the phase of the cell cycle where cells divide, and it’s also when RNAPII, the main transcription enzyme, is removed from the DNA. This suggests a direct link between the presence of active transcription machinery at TSSs and the delay in their replication.
G2/M DNA Synthesis: A Late Replication Opportunity
The study uncovered a significant phenomenon: a notable amount of DNA synthesis occurs in the G2/M phase, the period just before and during mitosis. This G2/M DNA synthesis, termed G-MiDS, is not associated with increased DNA damage, unlike mitotic DNA synthesis (MiDAS) which is often linked to DNA repair.
Further investigation showed that TSSs duplicated during G2/M possess specific characteristics. These TSSs often exhibit high levels of antisense transcription – transcription occurring in the opposite direction of the gene. This “antisense” transcription, along with other factors, seems to make these TSS regions challenging to replicate during the regular S phase, necessitating a later replication window in G2/M.
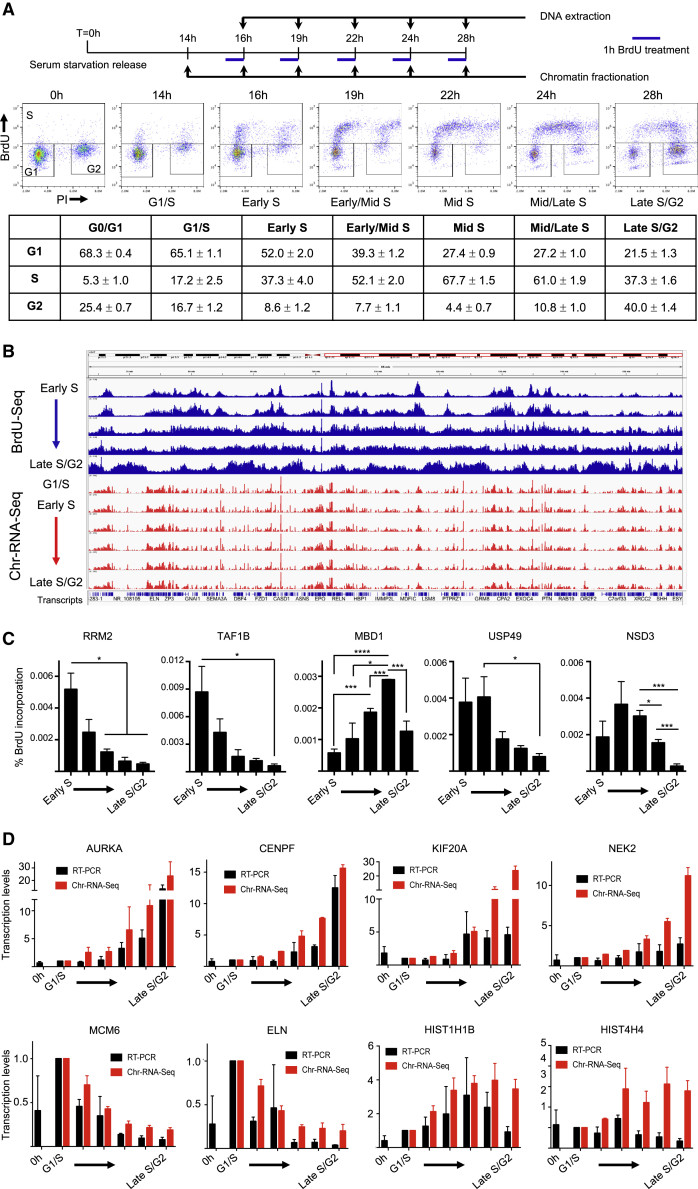 Experimental design and FACS analysis
Experimental design and FACS analysis
Implications of Delayed TSS Replication
These findings highlight a fascinating aspect of cellular coordination. While transcription and replication generally proceed simultaneously, active transcription, especially at TSSs, can create localized “roadblocks” for the replication machinery. The persistence of RNAPII and associated transcription factors at TSSs appears to impede efficient DNA duplication in these regions during S phase.
The cell cleverly resolves this conflict by deferring the complete replication of certain TSSs until G2/M, when the transcription machinery is temporarily disassembled during mitosis. This late replication ensures that even these challenging genomic sites are faithfully duplicated before cell division.
Asymmetric Replication Fork Progression and Transcription Conflicts
The researchers further explored the mechanisms behind TSS under-replication. By analyzing Okazaki fragments, short DNA pieces synthesized during replication, they found evidence of asymmetric replication fork progression near TSSs. Replication forks moving towards TSSs seemed to be less efficient, potentially stalled by the presence of the transcription machinery.
This suggests that while replication origins can be located near TSSs to initiate replication of transcribed genes, the replication fork encountering active transcription at the TSS experiences a slowdown, leading to the observed under-replication.
G-MiDS: A Distinct Process from DNA Damage Repair
Interestingly, G-MiDS, the G2/M DNA synthesis at TSSs, doesn’t appear to be a DNA damage response mechanism. It’s distinct from MiDAS, which is known to be involved in DNA repair during mitosis. G-MiDS occurs even in unperturbed cells and doesn’t rely on typical DNA damage repair pathways. This indicates that G-MiDS is a programmed process for completing replication at specific sites, rather than a rescue mechanism for stalled replication forks or damaged DNA.
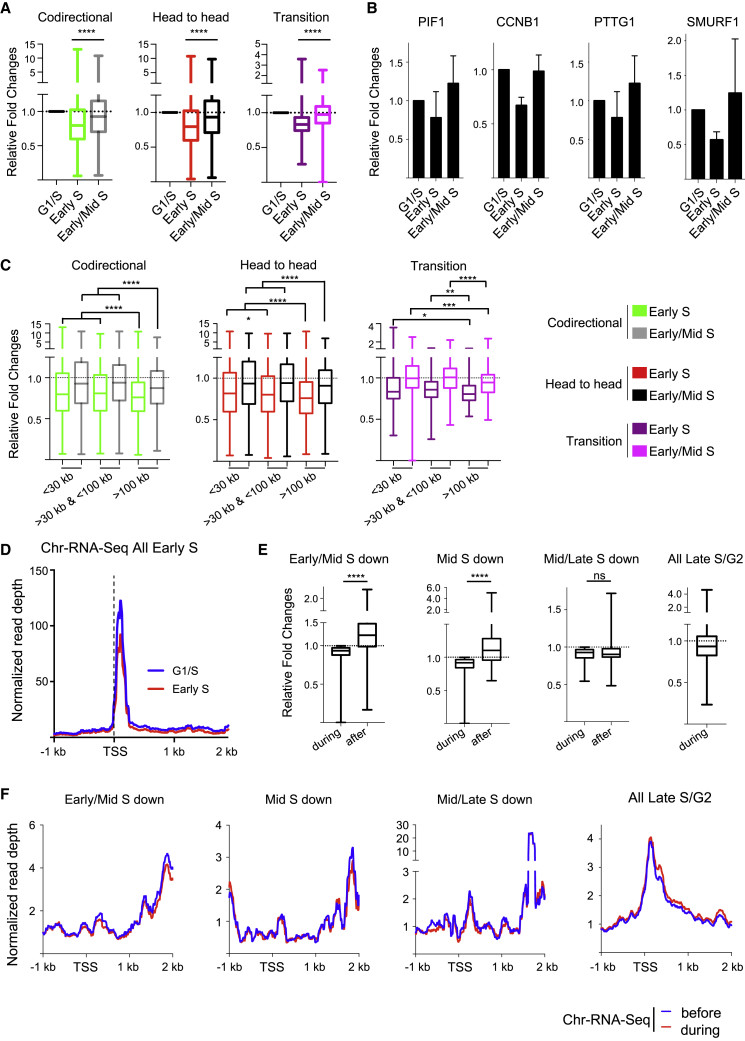 G-MiDS and replication timing
G-MiDS and replication timing
Features of G-MiDS Hotspot Genes
The study identified specific genes with TSSs that frequently undergo G-MiDS. These “hotspot” genes tend to be longer, highly transcribed, and enriched for antisense transcription. This combination of features likely makes their TSS regions particularly difficult to replicate during S phase, necessitating G2/M completion.
Many of these G-MiDS hotspot genes are involved in critical cellular functions, including transcription regulation, cell signaling, and cell migration. Intriguingly, some are also linked to cancer, raising the possibility that disruptions in G-MiDS and faithful replication of these TSSs could contribute to genome instability and disease development.
Conclusion: A Cell Cycle Orchestrated Dance
This research provides valuable insights into the dynamic interplay between transcription and DNA replication. It reveals that where transcription happens in relation to DNA replication is not uniform across the genome or throughout the cell cycle. TSSs of actively transcribed genes represent specific locations where replication is temporarily challenged by the ongoing transcription process.
The cell has evolved a sophisticated strategy to manage this conflict: transcription persists during replication, but replication completion at TSSs is often deferred to G2/M. This orchestrated delay, coupled with G-MiDS, ensures faithful genome duplication, even at transcriptionally active and structurally complex regions like TSSs, safeguarding genome stability and cellular function. Further research into the precise mechanisms and implications of G-MiDS, especially in the context of disease, promises to be a fruitful avenue for future discoveries.


